Why Structure Is Important
Think of a very large, randomly ordered list of numbers, and suppose you are searching for a particular number in that list with a computer. In order to find it, the computer will have to scan—in the worst case—the entire list of numbers. If instead those numbers were sorted, the computer would be able to perform binary search and only need to check a very small number of values, even if the original dataset was enormous. If the numbers are not in main memory, we could have adopted a B+-tree index and minimized the I/O operations needed to locate the desired number.
What the above example illustrates is that computers can analyze data much faster if the data is structured, which practically boils down to two things:
- Data layout: If the data values have some particular order on the storage medium, it is easier to search over them.
- Indexes: These are data structures built on top of the data values, which can expedite the search.
Database systems have been architected around data layouts and indexes to address performance when managing large quantities of data.
The market broadly categorizes data types as structured and unstructured. Unfortunately, structured data is tightly associated with tabular data (which is the data type on which the vast majority of database systems focus), dismissing anything non-tabular as “unstructured”. As a result, all complex, non-tabular data types typically end up in bespoke file formats and parsing tools, suffering from severe performance issues at the detriment of discovery.
At TileDB, we argue that no data is unstructured. We just need to unlock the structure in each data type, apply the appropriate data layout, and build the right indexes for efficient data processing. In the remainder of this section, you will dive deeper into the structure behind tabular data, how non-tabular data can also be structured, how to move away from bespoke file formats, and the power of a data structure called the multi-dimensional array, which is a first-class citizen in TileDB. The section concludes by affirming our thesis that no data is unstructured.
Tabular data
Tables and dataframes are universally considered structured, and for a good reason. Each table adheres to a specific schema that defines the columns and their data types. In addition, database systems typically impose certain row and columnar layouts, and build sophisticated indexes depending on which database operators they wish to boost. In addition, there can be support for data constraints within and across tables, as well as often strong consistency guarantees upon ingesting and querying the data.
Indeed, tabular data is very structured, and many solutions already exist in the market for efficiently managing tabular data. But what about anything non-tabular?
Non-tabular data
A broad misconception exists that all non-tabular data is unstructured. The most commonly used example is an image. You would avoid storing an image as a table. This is because you’d have to store each pixel in a table row in the form of (w, h, R, G, B, A), where w is the width coordinate of the pixel, h is the height coordinate of the pixel, and RGBA are the color values of the pixel. The addition of (w,h) in the physical storage, plus the way a SQL tabular query works, would absolutely destroy performance when accessing image data (especially if the image is huge, sometimes in the order of TBs for several applications). Indeed, tables would not be a natural fit for images, but is image data unstructured?
Image data is super structured! It consists of pixels laid out in a 2D matrix, where each element of the matrix has one or more color values, all of the same type across the color attribute. It’s in fact way simpler than a table. The key here is to understand that, although a table is not a good way in which you should lay out and index data, there may be other data structures that are more suitable for this data type (one of which we describe later in this section).
Similar arguments can be made for a wide spectrum of data types, such as video, audio, key-values, genomics, point clouds, and even text. Since all this data does not fit in tables, users resort to other means of storage: bespoke file formats.
Complex file formats
For any data type that does not naturally fit in a table, data analysts and scientists ended up creating their own structures in the form of bespoke file formats. Some file formats are easy (e.g., TIFF for imaging). Some are way more convoluted, such as in genomics and single-cell.
There several problems with bespoke file formats:
- No database features: Most bespoke file formats are accessible only by equally bespoke command-line tools, which lack important database features such as access control, logging, consistency guarantees and more. They are typically treated as plain files and managed by a file manager.
- Not cloud-optimized: Most of the file formats predate cloud object stores and, thus, have not been optimized for them. This means that users need to download potentially massive files from an object store to their local machine, even if they wish to slice just a small portion of this data.
- Resistant to change: Most of the file formats have been around for a very long time, and their specifications are defined by specific committees that decide on how and when to update them. Since potentially many tools are built on top of these formats, change can be very disruptive and is often avoided at all costs. Unfortunately, this means that these formats cannot take advantage of any technological advancements (such as cloud object stores), and the users try to depart from those formats by inventing more new, complex file formats.
So what can be a solution to this problem? We need to learn from history, and specifically from the successes on tabular data:
- Build databases, not formats: Similar to the way we created database systems around tabular data, we need to either evolve the tabular databases to handle non-tabular data too, or invent new ones from scratch.
- Standardize APIs, not format specs: Formats must always evolve. Therefore, we should not be looking to standardize the data layout, but rather the APIs to access the data and the functionality the users need. This is what we did with SQL for tables; two databases may adopt two different formats, but as long as they both comply with the same SQL standards, you can migrate from one to another without much disruption.
One question arises: is it possible to build a database that captures most—if not all—non-tabular data? It all boils down again to the two aspects of structure: data layout and indexes.
The power of arrays
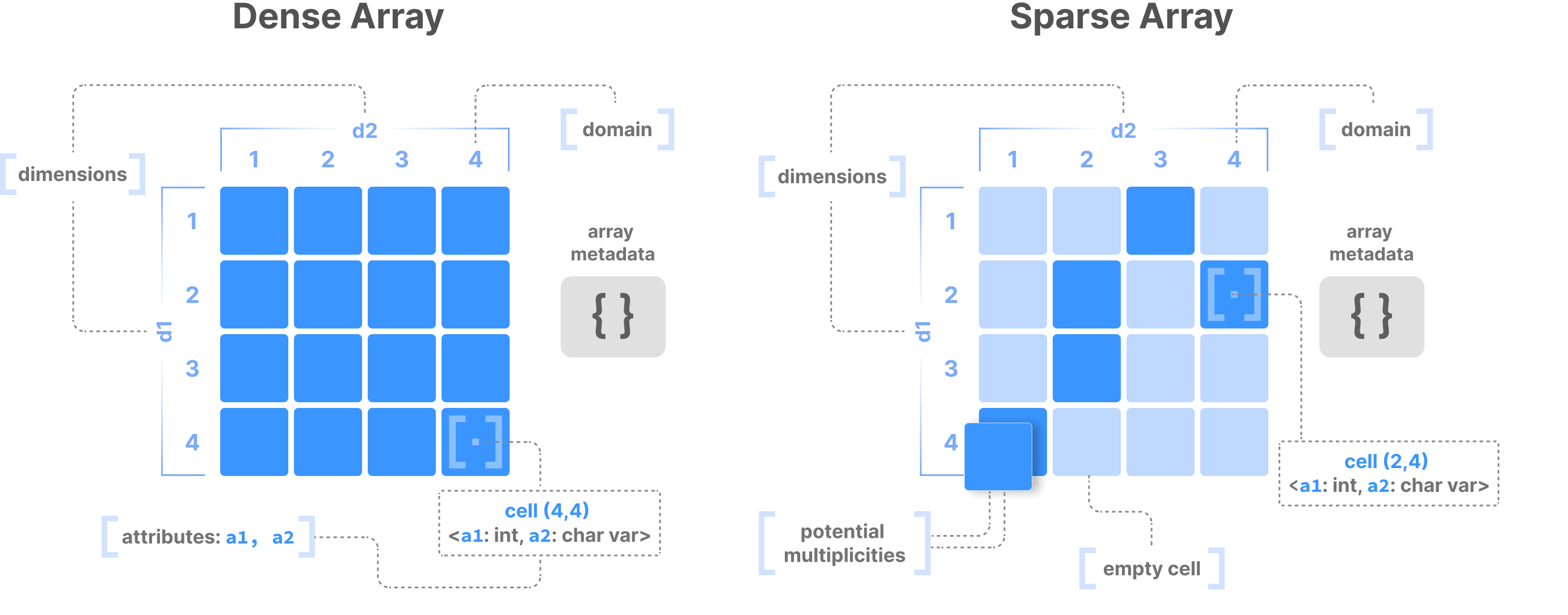
One data structure has consistently shown extreme promise when it comes to modeling diverse and complex data: the multi-dimensional array. The following figure depicts an example in the 2D case, making an important differentiation between dense and sparse arrays. A dense array contains values in all its cells, whereas the majority of the cells in a sparse array are empty, zero, or undefined. Array tiling flexibly defines the data layout, whereas the array dimensions collectively act as a multi-dimensional index that boosts search. Read the Array Data Model section for more details on arrays.
A table is just a special case for arrays, where one or more columns can be the array dimensions (which comprise the index), and the rest of the columns the array attributes. An array system can handle tabular functionality, and none of the aspects of tables prevent arrays from capturing them.
But things get more interesting in use cases where tables are not a good fit, such as images. An image is just a 2D dense array, where the RGBA values are just array attributes. The critical point with dense arrays is that they do not materialize the width and height coordinates of the pixels, offering close-to-optimal storage and slicing performance. That is not possible with tables.
Another good example is vector embeddings used in similarity search/nearest neighbor search and RAG LLMs in Generative AI. A vector is simply a 1D dense array, whereas vector search relies on indexes built on top of vectors for efficient nearest neighbor search. Taking it one step further, all flat files can be modeled as 1D dense arrays (a list of bytes).
Arrays can capture the majority of the data types we have seen in multiple industries: tables, images, point clouds, geometries, genomic variants, single-cell and spatial transcriptomics, video, audio, vector embeddings, flat files, and many more.
TileDB is architected around multi-dimensional arrays, which make it a multi-modal database, one that evolves way beyond tabular data.
No data is unstructured
We argue that, as long as humans understand a data type, it has structure behind it. The only truly unstructured data we can think of is white noise and ciphertexts. At TileDB, we are on a mission to help organizations discover insights from all their data, no matter how diverse and complex. As such, TileDB can handle all structured and “unstructured” data in one place, hoping to eventually convince everyone to remove “unstructured” from their vocabulary.