Spatial Data Model
The classical SOMA data model proves a natural representation for key elements of the multiplexed assay in the form of an expression matrix with cell and gene annotations. The spatial omics data model adds structure and new datatypes to the core SOMA data model to enable storing spatially resolved data in a way that captures the underlying structure of the spatial context. For a refresher on the SOMA data model, refer to the SOMA Data Model.
SOMA groups spatial objects together based on their presence in a single physical space by introducing the notion of a scene. A scene is a collection of spatially resolved information defined on a single coordinate space. The scene stores coordinate transformations from the elements stored in it back to the shared coordinate space. This allows querying and analyzing different elements within the same physical space together.
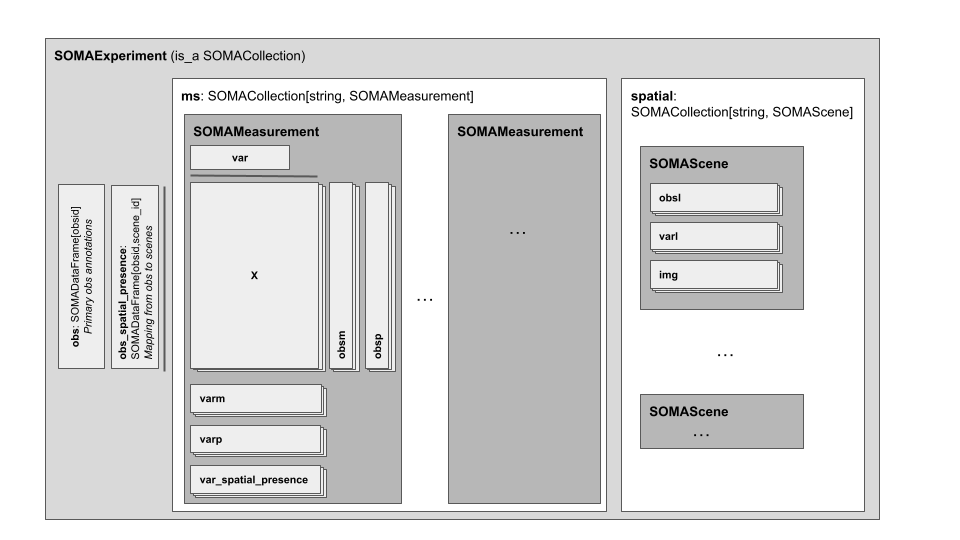
As before the SOMAExperiment is the top-level, multi-modal container for a TileDB-SOMA dataset. It includes the new elements:
spatial, aSOMACollectionthat stores one or moreSOMASceneobjects.obs_spatial_presence, an optionalSOMADataFramethat stores the presence of each observation in each scene.
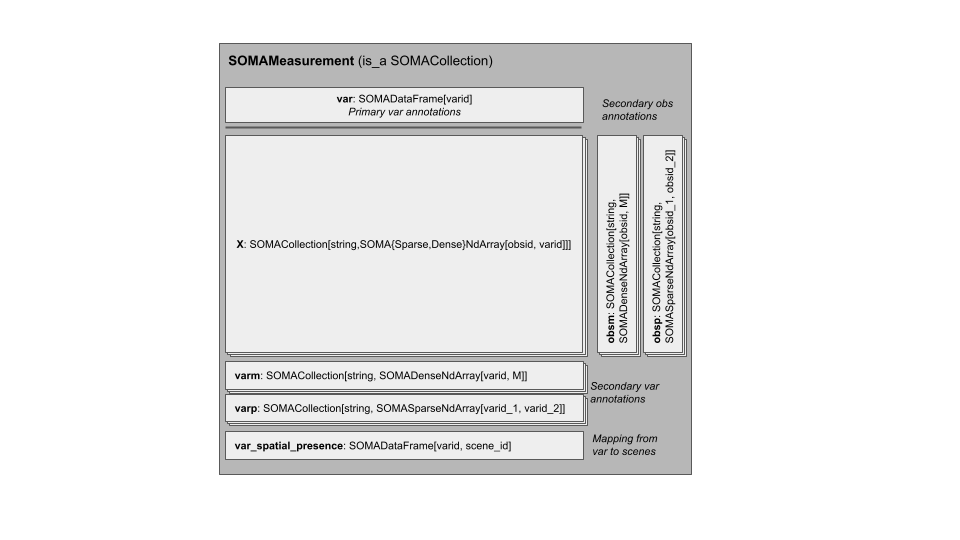
The SOMAMeasurement is still the container for a single modality (for example, gene or protein expression) for the set of observations in the experiment. It now includes a new element for cataloging spatial relationships:
var_spatial_presence, an optionalSOMADataFramethat stores the presence of each variable in each scene.
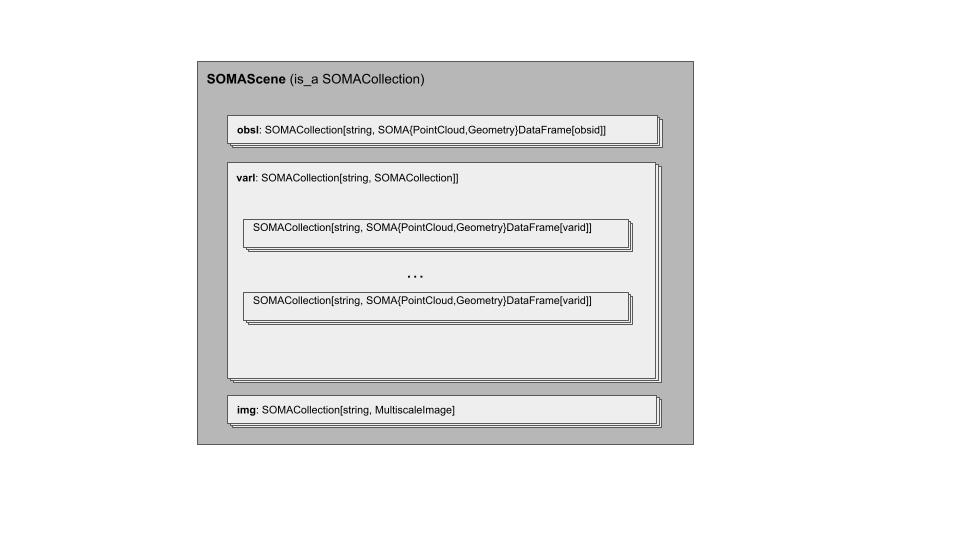

The SOMAScene is a new container for spatially resolved data (for example, spot locations of observations or tissue images). The SOMAScene has a coordinate space, and elements within the SOMAScene have transformations that map them to that shared coordinate space. It has the following elements:
img, aSOMACollectionofSOMAMultiscaleImagesrelated to the experiment.obsl, aSOMACollectionofSOMAPointCloudDataFramesandSOMAGeometryDataFrames. These store location-based annotations on the observable domain. Thesoma_joinidin any item in this collection should be interpreted as theobsid.varl, aSOMACollectionofSOMACollections, each of which are collections of one or moreSOMAPointCloudDataFramesorSOMAGeometryDataFrames. These store location-based annotations on the variable domain. The outer collection is keyed on the measurement names. Thesoma_joinidfor items in the inner collection should be interpreted as thevarid.
For more technical details about SOMA’s data structures and data model, visit the [SOMA specification][spec].