import os
import shutil
from tiledb.bioimg.converters.ome_tiff import OMETiffConverter
from tiledb.bioimg.openslide import TileDBOpenSlide
root_dir = os.path.expanduser("~/tiledb-bioimg-openslide")
if os.path.exists(root_dir):
shutil.rmtree(root_dir)
os.makedirs(root_dir)Inspect Biomedical Images with the OpenSlide API
Slicing and reading your data in TileDB is an essential step to accessing and deriving value from your data. For this reason, you have two options on how you can read your data into TileDB. In this tutorial, you will learn how to read and slice your biomedical imaging data by using the TileDBOpenSlide API. This API has been designed on top of the popular OpenSlide API, to offer a familiar means to access your data.
Setup
Start by importing the necessary modules for this tutorial and creating a directory to hold the data.
Image properties
Start by ingesting an image into TileDB.
You can create a slide of the image by using the TileDBOpenSlide wrapper and query the image properties as follows:
The API offers also a more granular access to each level’s properties, where you can query information about each resolution layer
Read a resolution level
You can read a whole resolution level by defining the level ID:
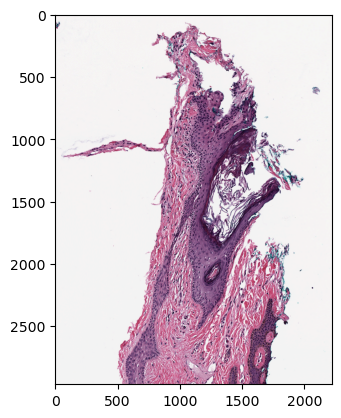
You can then visualize it as follows:
Slice a resolution level
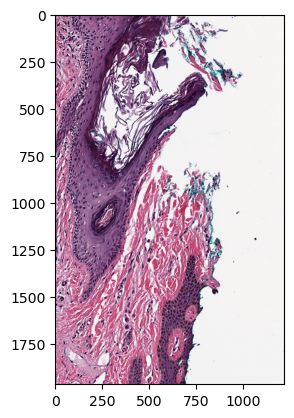
You can slice image data of a specific level with the read_region method by defining the upper left coordinates and the size of your slice.