Usage
This section provides a brief overview of the fundamental concepts of notebooks within TileDB. For a comprehensive introduction to the Jupyter environment and project, please visit the Getting Started materials from Project Jupyter.
Cell-based workflow
A notebook is composed of cells, each of which may contain code, text, or other content, presented based on the cell type. Cells within a notebook may be edited, run, re-edited, and re-run repeatedly during the analysis process in order to reach a desired program state.
Kernels
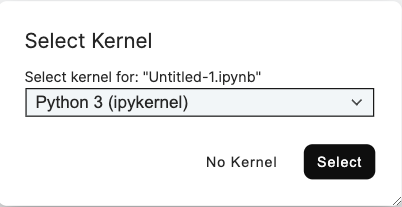
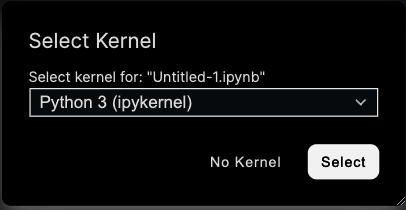
For code execution, each notebook is connected to a kernel, which is a separate Python or R process running on the TileDB notebook server. This process is independent of the notebook interface, and kernels may be restarted without losing work within the notebook itself. A kernel must be selected when a new notebook is created within the notebook user interface.
Code cells
The default cell type in a notebook is a code cell, which may contain code written in Python, R, or potentially other languages if a suitable kernel is configured. The language used within a code cell must match the selected kernel for the active notebook. All language-specific syntax and formatting rules apply to code within a cell. For example, Python code must be properly indented and follow all other significant whitespace rules.
Cells are most commonly run one at a time, after entering or updating code within the cell. To send the code to the active kernel for execution, use the “play” (right-arrow) button in the notebook menubar, or press Shift+Enter.
After execution, the output of the given code will be presented in a separate display box below the code cell. The contents of this box may be text output or a plot generated by the program, or even interactive widgets. With the exception of widgets, most output cannot be modified, but it can be selected and copied.
For shared notebooks, it is best practice to restart the kernel and re-run the notebook top-to-bottom before saving, in order to make sure that all inputs and outputs still match expectation when run consecutively rather than ad-hoc while editing.
Markdown cells
Alternatively, a cell may contain Markdown-formatted text. Markdown cells are used for explanatory structured text, which requires styling for readability. To create a Markdown cell, use the cell type selector box:
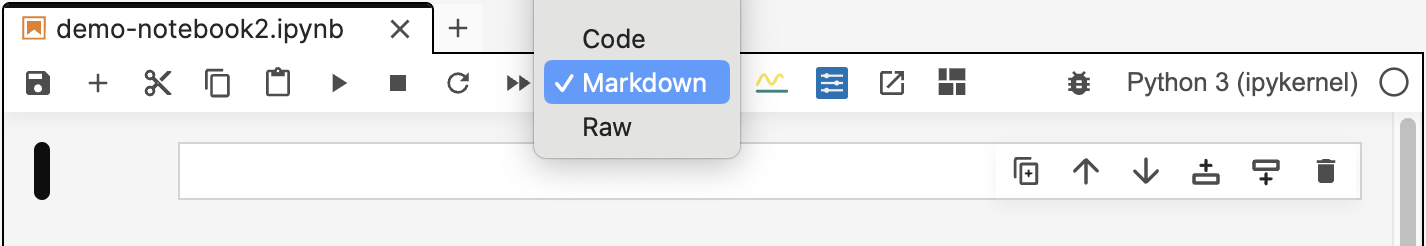
And choose Markdown as the type:
While you may “run” these cells in the same manner as shown above, the content is never actually executed by the kernel. Instead, Markdown cells are rendered into a more presentable format, which is displayed instead of raw text, until the cell is edited again. For example, the following text shows several features of Markdown:
# Hello, world! This is a heading.
## Sub-heading
This text is within a sub-heading.
### Sub-sub-heading
The following text is _italicized_, and now also **_italicized and bold_**.Which will be rendered as follows:
Install packages
TileDB notebook servers include a persistent home directory for you to install custom packages. TileDB supports pip, conda/mamba, CRAN, and other methods of installing packages. When installing a package, you should install it in your home directory if you want the package to persist after reboots.
For Python, you can use pip to install packages in your home directory with the --user option.
pip install --user <package>If you install a package from pip without the --user flag, it will not be persisted and will not be available upon reboot.
Both conda and mamba are available in the notebook environment to install any available packages. By creating a custom environment in your home directory, you can install and persist any packages.
mamba create -n my_env
mamba activate my_env
mamba install -c conda-forge <package>If you use conda install or mamba install outside of a custom environment, your packages will not be persisted.
You can install R packages from CRAN by setting the lib argument to the location to the home directory for persistence.
install.packages("package_name", lib="~/R")If you install packages without setting the library path to your home directory, it’s likely the installed packages will not be persisted.