Genomics (VCF)
TileDB architects its entire data engine around the multi-dimensional array, a powerful data structure that shape-shifts to adapt and structure any modality, no matter how complex. VCF datasets are able to take advantage of the multi-dimensional array format and are made up of 3-dimensional sparse arrays. You can learn more about VCF datasets and population genomics at Structure: Population Genomics.
Add VCF
TileDB Cloud provides a method to ingest a batch of single sample VCF files into a VCF dataset and add the dataset to the catalog, all in one step. The source VCF files are read from cloud object store and written into TileDB arrays defined by TileDB-VCF. You can learn about the mechanics, theoretical background, and detailed tutorials in section Structure: Population Genomics.
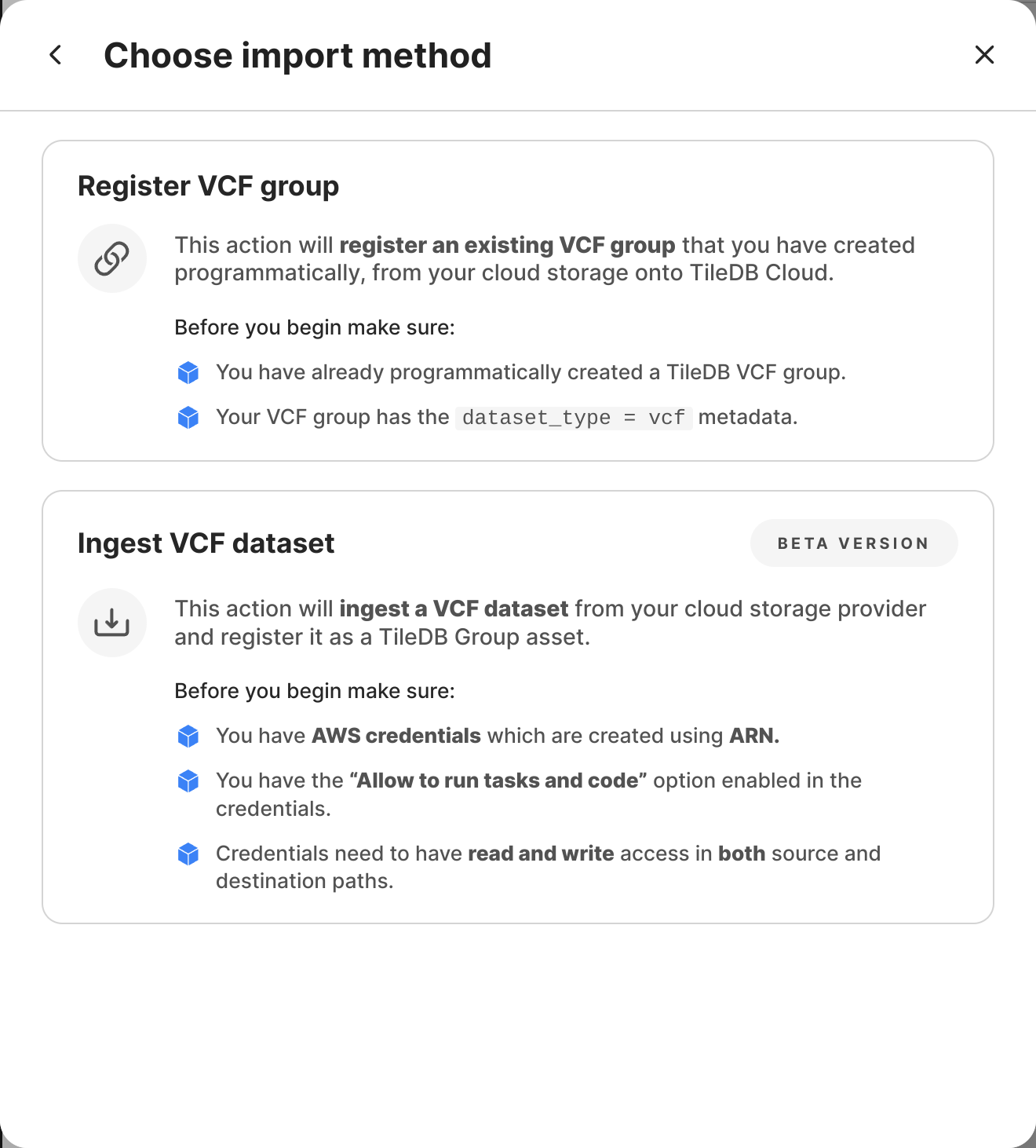
You can add a VCF dataset to the TileDB catalog in one of two ways:
- Create and register in one step: TileDB offers you a means of one-click ingestion in the TileDB UI.
- Create first, register after: Alternatively, you may first physically create a VCF dataset in your object store, without necessitating cataloging it with TileDB as well. At any point in time, you can register any existing VCF files with TileDB using the UI or an API command.
Create and register in one step
To ingest a batch of VCF files into a VCF dataset, perform the following steps:
Navigate to the Assets section.
Select Add asset.
Select Data as the type of asset to add.
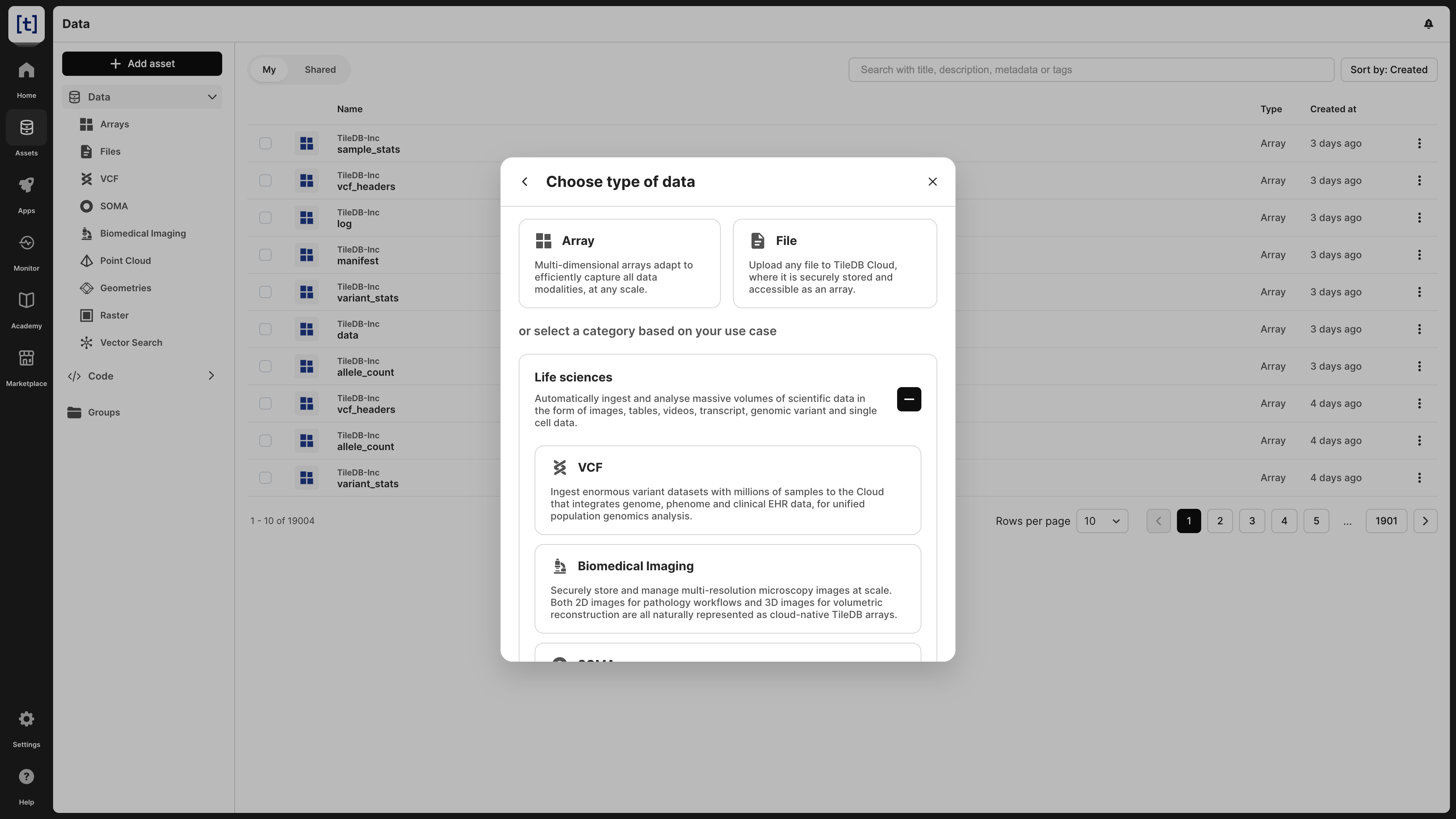
Expand the Life sciences category and select VCF.
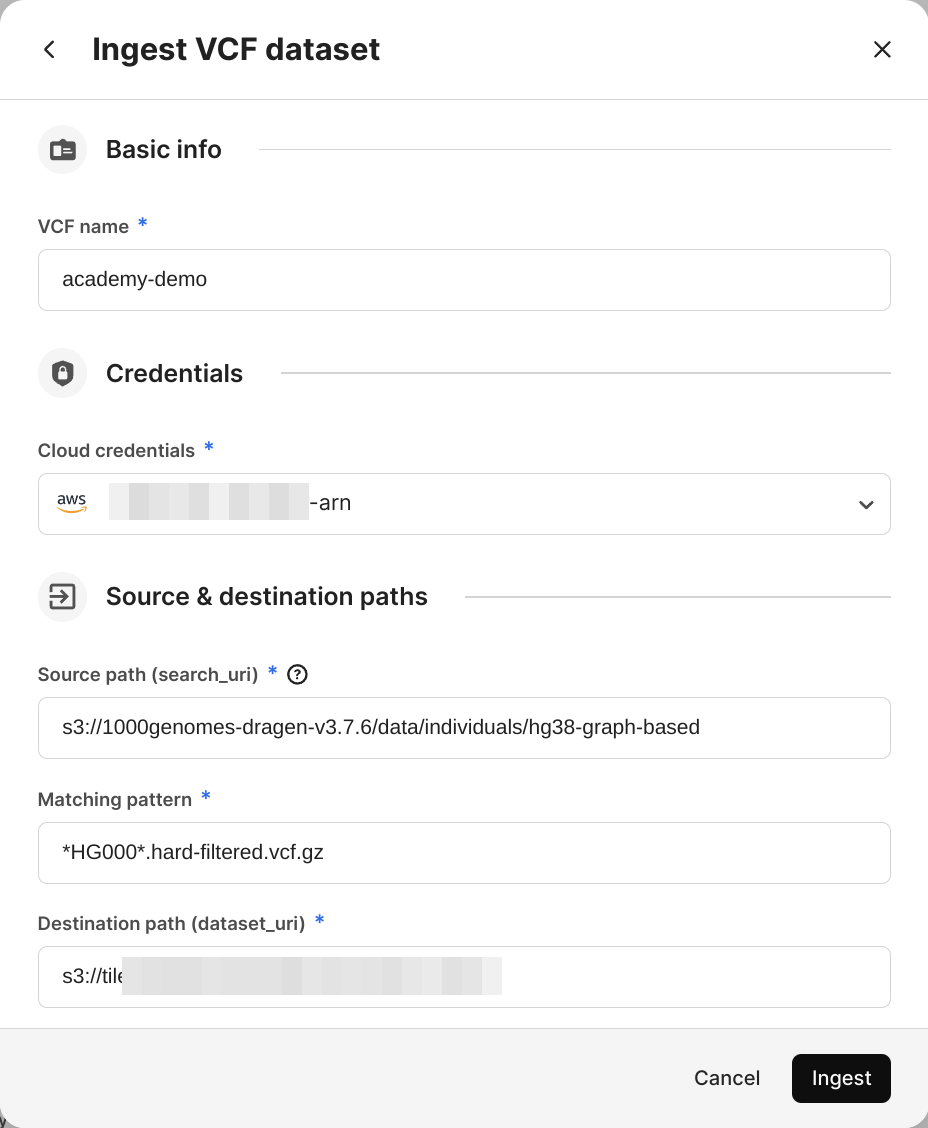
Select Ingest VCF dataset.
Add a VCF name of your choice, which will be the VCF dataset asset name on TileDB Cloud.
Select the Cloud credentials used to access cloud storage.
Add a Source path, which is the cloud storage URI that will be recursively searched.
Add a Matching pattern, which is a glob pattern used to select the VCF files being ingested.
Select Ingest.
The ingestion task graphs can be viewed in the Logs - Task graphs monitor.
Once the ingestion completes, you can view the VCF dataset in Assets.
For steps on how to programmatically create and register a VCF dataset in TileDB, review the Population Genomics: Basic TileDB Cloud tutorial.
Create first, register later
Assuming you have already created a VCF dataset and stored it in S3, perform the following steps to register it in the UI:
Navigate to the Assets section.
Select Add asset.
Select Data as the type of asset to add.
Expand the Life sciences category and select VCF.
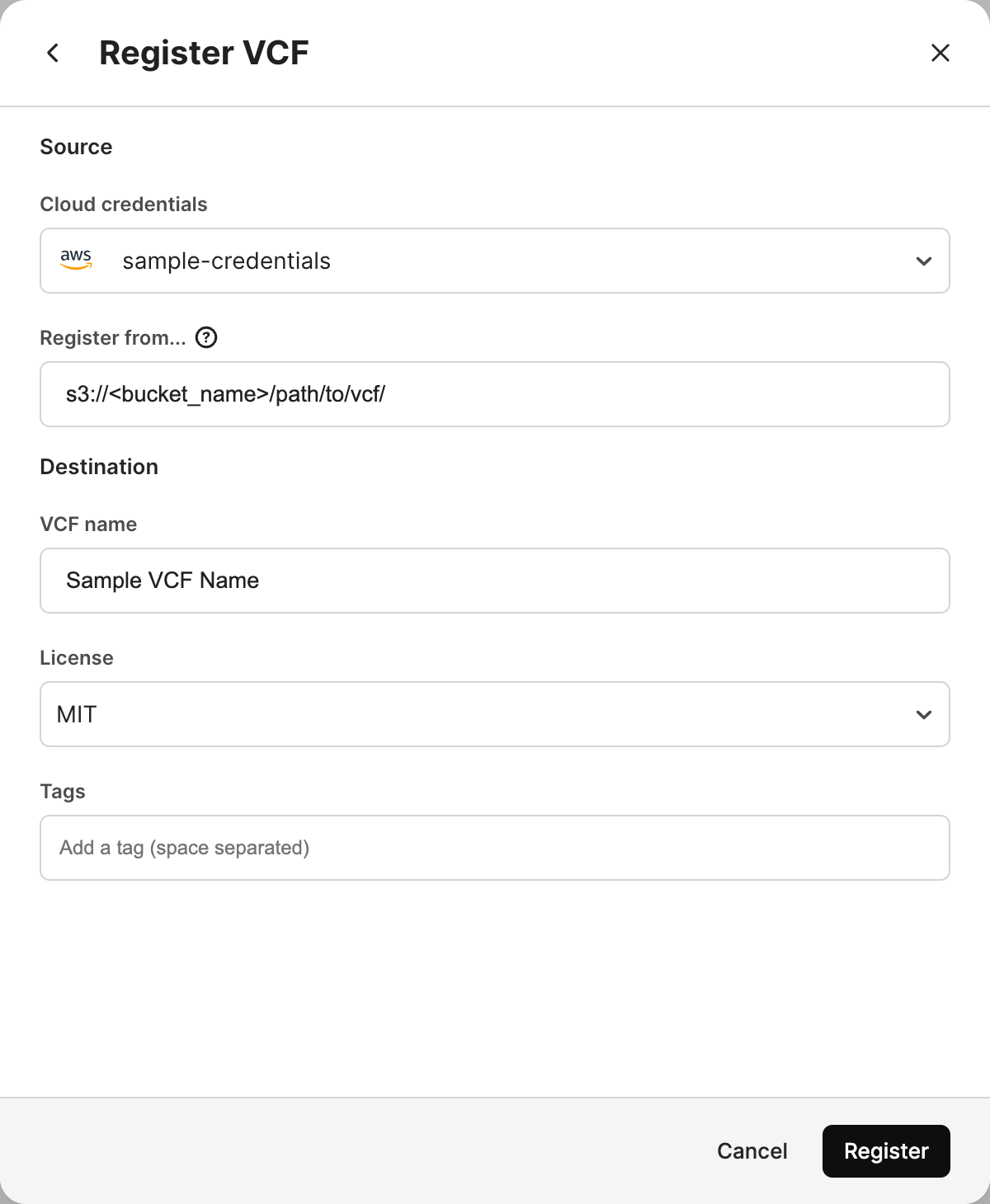
Select Register VCF group.
Choose the appropriate Cloud credentials to access the VCF.
Specify the URI where the VCF dataset lives in the Register from… field. This is the full URI including the VCF dataset folder.
Specify a meaningful VCF Name.
Optionally specify a License for your VCF dataset. This is especially important if you make your VCF dataset public.
Optionally specify Tags for your VCF dataset.
Select Register.
Overview
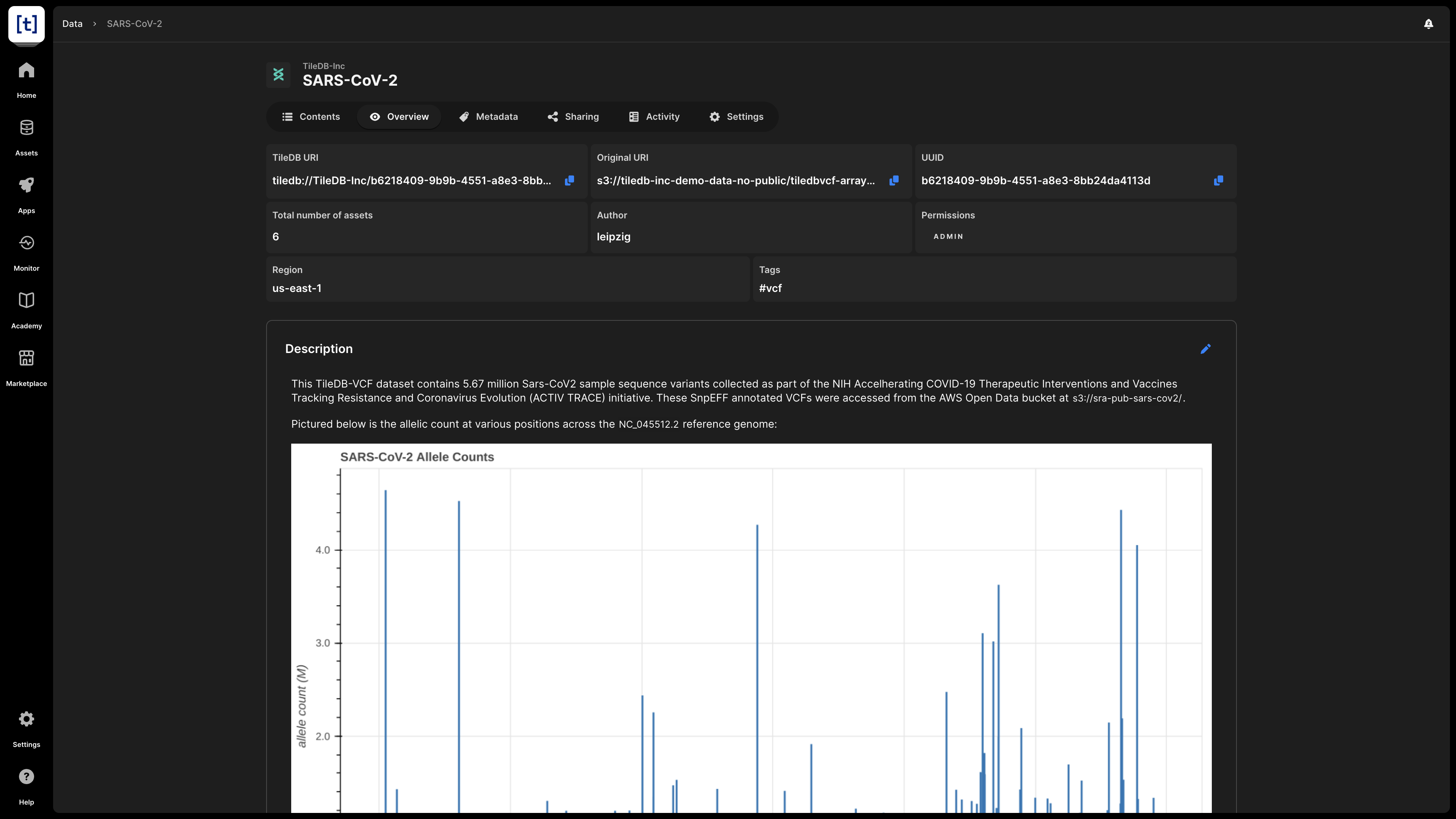
In this screen, you can find basic information about the VCF dataset:
- VCF name - This appears at the very top of the screen, and consists of the account name and the name you provided to the VCF dataset when you registered it.
- TileDB URI - The unique resource identifier for TileDB, based on which you can call the VCF dataset when coding. It comprises the namespace identifier and the UUID of the VCF dataset.
- Original URI - The location on cloud storage where the VCF dataset is physically stored.
- UUID - The unique identifier for the VCF dataset.
- Total number of assets - The total number of assets, which you can preview in the Contents tab.
- Author - The author of the asset.
- Permissions - What rights the current user has on this VCF dataset. Possible values are
READandADMIN. - Region - The region in which the VCF dataset is stored on cloud storage.
- Tags - Any tags attached to the VCF dataset for searchability purposes.
- Description - If the user has provided a description to the VCF dataset (programmatically or in Settings), it is visible here. The description is indexed and searchable in the catalog. Therefore, it’s recommended to add a meaningful description for all your assets.
It is important to understand how to refer to your VCF dataset programmatically. You can do it in two ways:
- Using the TileDB URI format
tiledb://<account>/<vcf_name>. This is the most user-friendly way, but TileDB allows duplicated VCF dataset names, and if you have a VCF dataset with a non-unique name, this will throw an error. - Using the TileDB URI from the asset’s Overview tab (that is, the URI with format
tiledb://<account>/<UUID>). TileDB URIs referencing the asset’s UUID are unique. Thus, this method will always work.
You can programmatically get overview information about the VCF dataset with the following command:
# The following will return a JSON file with various info about the VCF dataset.
tiledb.cloud.asset.info("tiledb://<account>/<vcf_name>")Contents
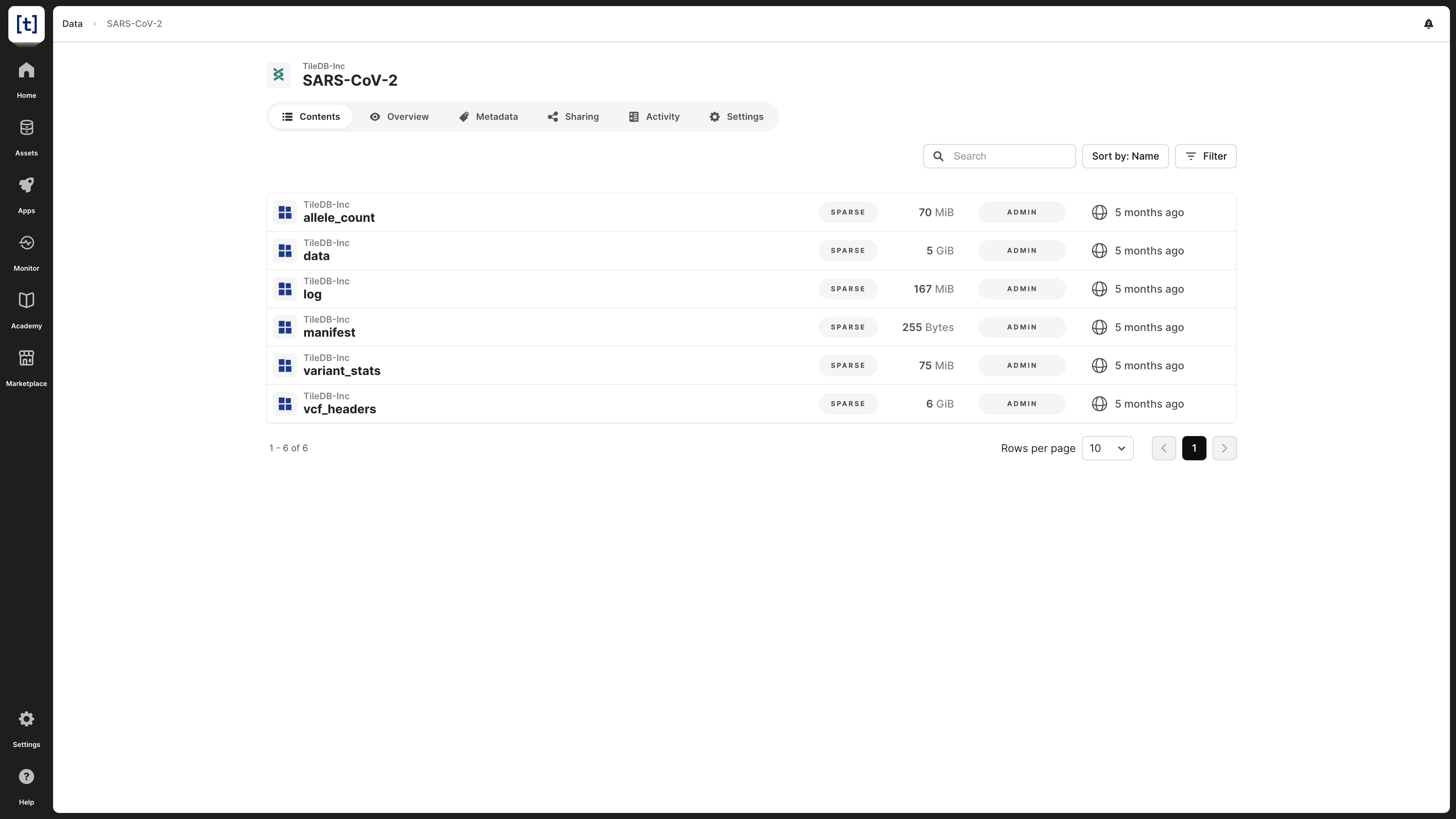
In the Contents tab of a VCF dataset, you can see all of the assets that comprise the VCF dataset, along with their sizes, your permissions on those assets, and when the assets were last modified. You can also select one of the assets to view details about the asset in the catalog.
For information about the purpose of each asset in a VCF dataset, visit Population Genomics: Data Model.
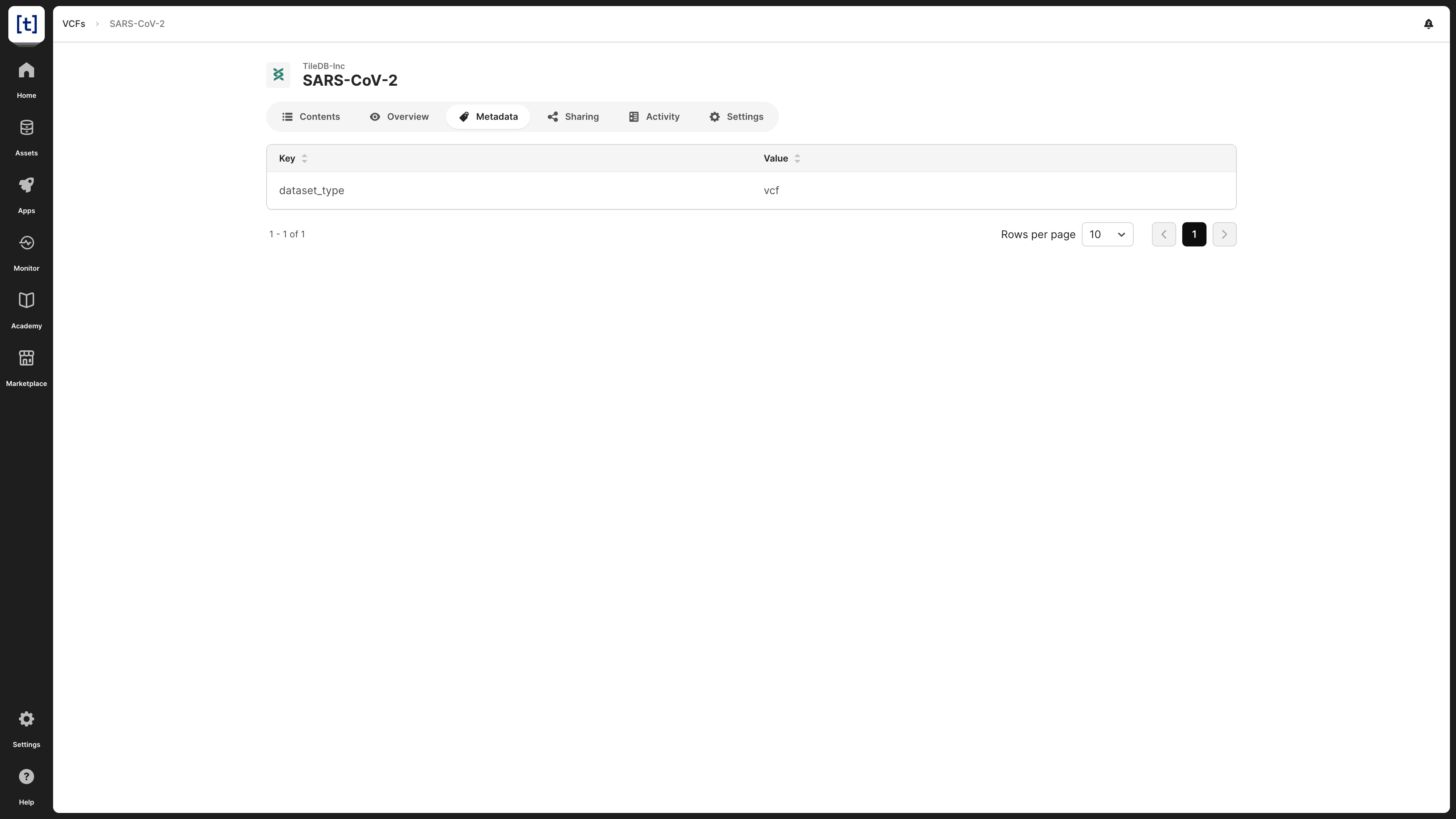
Metadata
VCF datasets may be associated with metadata in the form of key-value pairs, which is visible in the Metadata tab.
Settings
In the VCF dataset settings, you can:
- Add a description - Note that this is indexed and, thus, searchable in the TileDB catalog.
- Mark the VCF as read-only - This is useful if you want to prevent any changes to the VCF dataset by you or someone with whom you shared the VCF dataset.
- Make public - If you wish to share the VCF dataset with all TileDB users. This will appear in the Marketplace tab in the left navigation menu. If you make a VCF dataset public, you can easily change it back to private in the same manner.
- Change cloud credentials - Credentials should be provided so that TileDB can securely access the VCF on the cloud store where it is physically stored.
- Rename VCF - Take caution when renaming VCF datasets, as any URLs including the previous VCF dataset name will no longer work.
- Delete VCF - Visit Delete VCF for more information.
You can programmatically update some VCF dataset settings with the following command:
tiledb.cloud.asset.update_info(
uri="tiledb://<account>/<vcf_name>",
description=None, # Optional - A new description
name=None, # Optional - A new name for the VCF dataset
tags=None, # Optional - VCF dataset tags that will be searchable in the catalog
credentials_name=None, # Optional - The cloud credentials that access the VCF dataset (should already exist in your account settings)
)To make a VCF dataset public programmatically, run the following:
tiledb.cloud.asset.share(
"tiledb://<account>/<vcf_name>", namespace="public", permissions="read"
)Rename VCF
A useful property of the TileDB catalog and the way it registers VCF datasets is that you can easily rename a VCF dataset, without physically moving it, thus avoiding the very expensive copying operations entailed in object stores when physically renaming/moving file objects. You can rename VCF datasets from the Settings tab.
You can programmatically rename a VCF dataset as follows:
tiledb.cloud.asset.update_info(
"`tiledb://<account>/<previous_name>`", name="<new_name>"
)Take caution when renaming VCF datasets, as any URLs including the previous VCF dataset name will no longer work.
Delete VCF
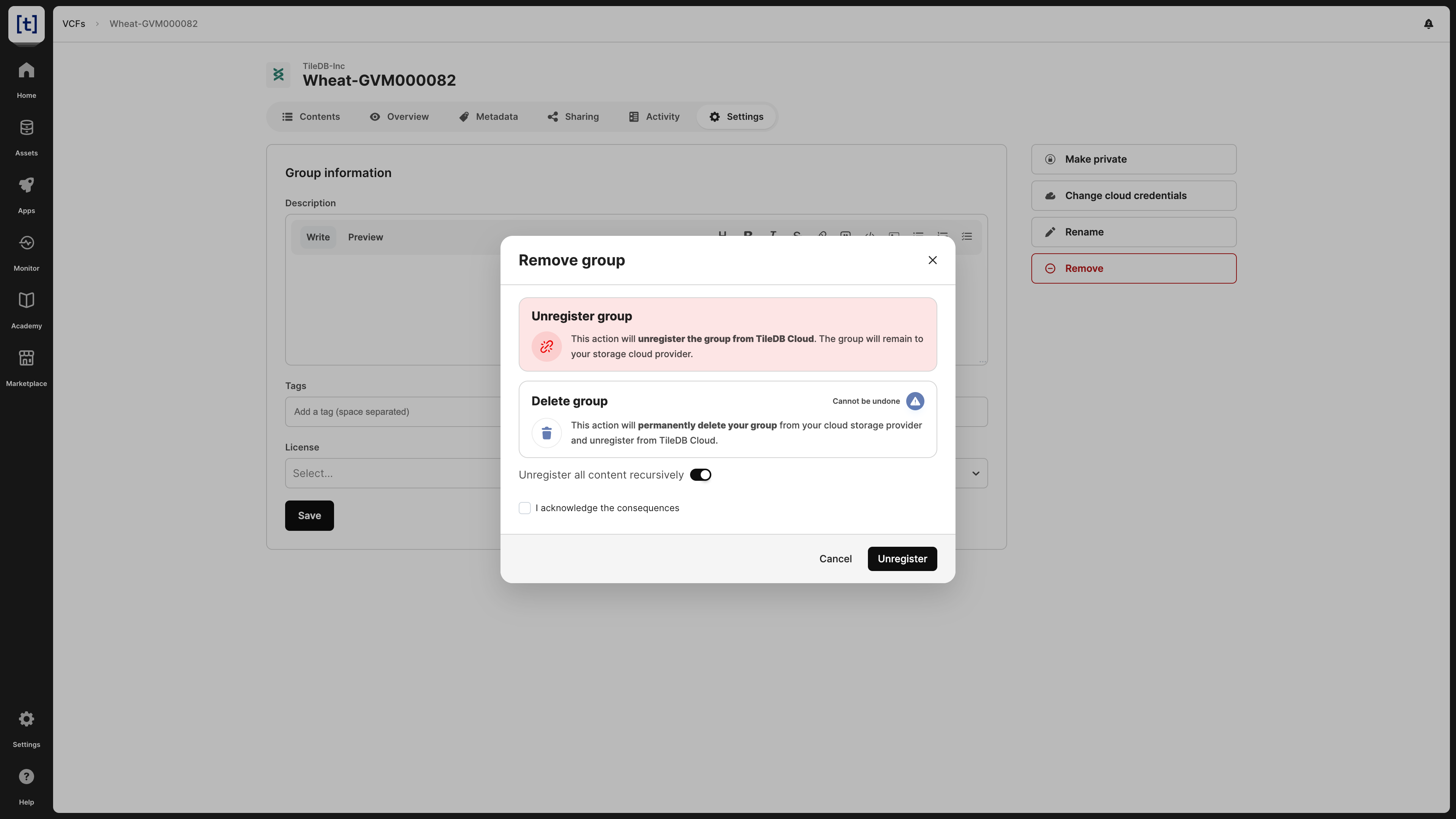
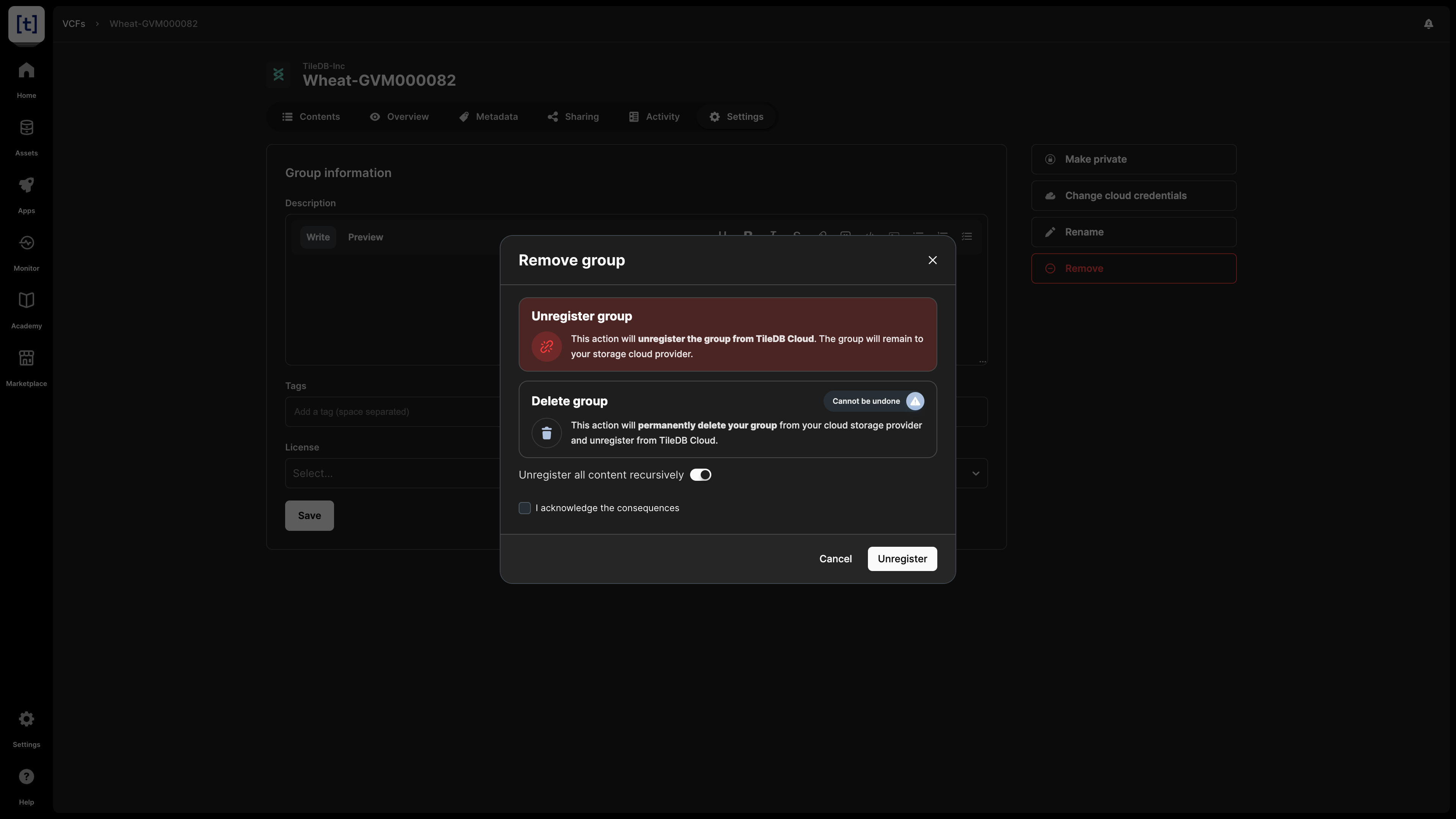
When deleting a VCF dataset, you have two options:
- Unregister: This operation removes the VCF dataset from the TileDB catalog, but it does not physically remove it from the object store. Since the VCF dataset will persist on storage, you can register it again in the TileDB catalog in the future.
- Delete: This operation both unregisters and physically removes the VCF dataset from storage. Note that this operation cannot be undone.
For both unregistering and deleting a VCF dataset, you have the option to apply the operation recursively. Unregistering a VCF dataset recursively involves unregistering both the VCF dataset and the arrays contained in the VCF dataset from TileDB, but neither the VCF dataset nor the arrays contained in the VCF dataset are removed from your cloud object store. Deleting a VCF dataset recursively deletes both the VCF dataset and the arrays contained within the VCF dataset. Deleting a VCF dataset is permanent and cannot be undone.
You can delete the VCF dataset from the Settings tab, which will prompt you to choose among the two operations above.
You can also programmatically delete or unregister the VCF dataset as follows:
# Unregister a VCF dataset
tiledb.cloud.asset.deregister(uri="tiledb://<account>/<vcf_name>")
# Delete a VCF dataset
tiledb.cloud.asset.delete(uri="tiledb://<account>/<vcf_name>")