import matplotlib.pyplot as plt
import pyarrow as pa
import seaborn as sns
import tiledb.cloud
import tiledbsomaUse Distributed Compute with SOMA Datasets
You can run this tutorial only on TileDB Cloud. However, TileDB Cloud has a free tier. We strongly recommend that you sign up and run everything there, as that requires no installations or deployment.
TileDB Cloud’s serverless compute platform enables the execution of distributed compute tasks on single-cell datasets stored in TileDB-SOMA. This tutorial will cover how to:
- Run an arbitrary function serverlessly on TileDB Cloud as a user-defined function (UDF).
- Register a UDF with TileDB Cloud to simplify reuse and sharing.
- Build and execute a task graph composed of multiple UDFs to perform a distributed operation.
- Monitor the task graph’s progress.
- Retrieve and visualize the task graph’s results.
Background
Serverless? UDFs? Task graphs? Oh my! Here’s a quick primer in case these concepts are new to you:
Serverless: Serverless computing allows you to run code without having to provision or manager servers. Instead, TileDB Cloud automatically spins-up a cloud-based server to run your code, and then shuts it down when the code is done executing. That way, you only pay for the time your code is running, and you don’t have to worry about managing servers.
UDFs: User-defined functions (UDFs) are custom functions that you can write in a variety of languages and run on TileDB Cloud.
Task graph: A task graph is a directed acyclic graph (DAG) that represents a series of tasks (i.e., UDFs) that need to be executed in a specific order that also accounts for dependencies between them.
Objective
Your mission is to build a reproducible workflow for calculating gene-wise summary statistics for a user-defined set of genes across a collection of single-cell RNA-seq experiments. In this tutorial, you will work with a collection of 24 tissue-specific datasets produced by the Tabula Sapiens consortium. Each of these datasets has been converted to a SOMA experiment and published as a collection on TileDB Cloud.
Setup
You will need to import the tiledbsoma and tiledb.cloud packages to run this tutorial, as well as a few other common libraries for data manipulation and visualization.
Now define a couple constants that will be used throughout the tutorial.
NAMESPACE: The TileDB Cloud namespace that you are working in. You should update this with your own namespace.SOCO_URI: The TileDB Cloud URI for accessing the collection of Tabula Sapiens SOMA experiments.
Retrieve the list of SOMA experiments from the Tabula Sapiens collection.
Base function
The first step is to write the function that will calculate summary statistics for a given set of genes in a SOMA experiment. Then step-by-step you will see how to leverage TileDB Cloud’s functionality to run this function serverlessly and then in parallel across multiple experiments at once.
Now run it locally to verify that the function works as expected.
Perfect! Now, you will see how to serverlessly execute this function on TileDB Cloud.
Serverless execution
TileDB Cloud’s serverless compute platform allows you to run any UDF on the cloud. In practice, this means the function’s code is serialized and shipped to a cloud-based server where it is executed before the results are returned to your session.
Just pass the callable function and its arguments to tiledb.cloud.udf.exec() to execute it on TileDB Cloud.
As expected, the results are the same as when you ran the function locally. However, this time a TileDB Cloud-based server performed the operation, rather than the machine running this notebook.
This serverless approach provides several immediate benefits:
- Scalability: With very little effort, you can run any function on a server with more resources than your local machine.
- Cost: You only pay for the time the function is running, rather than having to maintain a server that is always on or go through the hassle of spinning up and shutting down servers as needed.
- Auditability: All serverless executions are logged and can be reviewed at any time. Visit https://cloud.tiledb.com/monitor/logs/tasks to see a list of all UDFs that have been executed, including the one you just ran.
Selecting the executed UDF’s entry in the TileDB Cloud Logs view will provide details about the UDF, including the function’s source code, how long it took to run, and the total cost of running it.
This information is also retrievable programmatically:
Learn more about TileDB Cloud serverless UDFs here.
Cloud server resources
By default, TileDB Cloud allocates 2 CPUs and 2 GB of memory to the server running your UDF. However, you can opt for a larger server with 8 CPUs and 8GB of memory by changing the resource_class argument from "standard" (the default), to "large".
For even more flexibility over the server’s resources, you can choose to execute the UDF in batch mode. Unlike realtime mode (the default mode), where the UDF is executed on a virtualized server that’s already running, batch mode execution spins-up a dedicated server to run your UDF. This provides full flexibility to specify the server’s CPU count and available memory, at the cost of a longer startup time. Batch mode is beyond the scope of this tutorial, but you can learn more about it in the Task Graph Modes section of Academy.
UDF registration
If you plan to reuse a UDF, you can register it with TileDB Cloud for easy reference in future executions.
This adds the UDF as a new asset in your TileDB Cloud data catalog, which can be viewed at https://cloud.tiledb.com/assets/udfs/my. Selecting the new soma_gene_stats UDF will show you the function’s source code, execution history, and other details. Now you can invoke the UDF by name (in the form of <namespace>/<udf_name>):
Just like any other TileDB Cloud asset, a registered UDF can be shared with collaborators or made public. Anyone with access to the UDF can execute it themselves without rewriting the function (you will not be charged when others run it).
Scaling with task graphs
Now it’s time focus on the mission at hand: calculating these summary stats for each of the tissue-specific datasets in the Tabula Sapiens project. Of course, you could run the soma_gene_stats() function 24 times in a loop, but who has the time for that? Running these operations in parallel will be much faster. And that’s where task graphs come in.
Task graphs are a powerful feature of TileDB Cloud that allow you to orchestrate the execution of multiple UDFs while taking into account dependencies between them. This is particularly useful when you need to run a series of tasks in a specific order, or want to parallelize the execution of a distributable task (or both), as is the case here.
Here, you will build a task graph that runs the soma_gene_stats() function on each tissue-specific SOMA experiment in parallel. The first step is to initialize a new task graph, specifying the namespace to charge and a name for the task graph.
Next, add the individual tasks to the task graph, one for each of the tissue-specific SOMA experiments in the collection. The easiest way to do this is to use a loop to submit a task for each of the experiment URIs in the exp_uris dictionary. Note that each iteration of the loop calls submit() with the same parameters, except for the experiment uri and name, which are unique to each SOMA experiment.
The .visualize() method offers a clear view of the task graph’s structure, so you can monitor the task graph’s progress. Each node represents a UDF, hovering over a node will show the UDF’s name and current status (for example, Not Started, Running, Completed, or Failed). Because all tasks are independent of each other, they will all run in parallel.
To kick off execution call compute() on the graph object. The wait() method will block the notebook until all tasks have completed.
You can monitor the progress of the task graph in real-time by visiting https://cloud.tiledb.com/monitor/logs/taskgraphs and selecting the task graph you just executed.
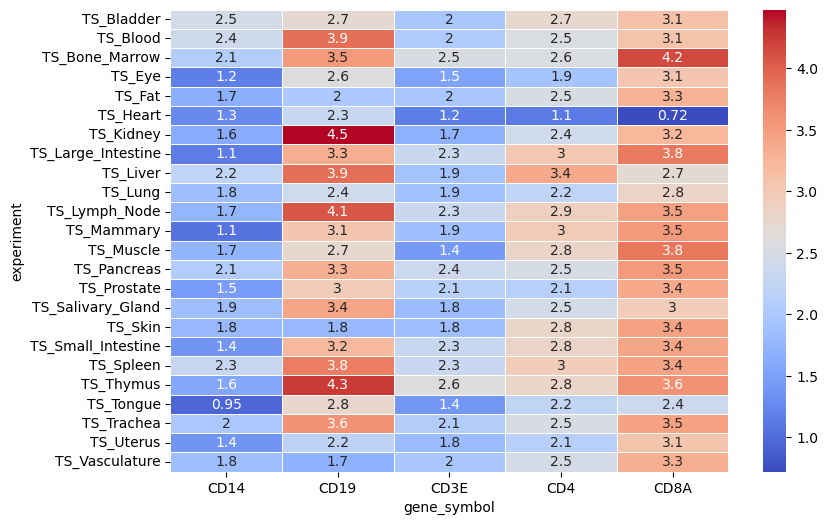
Retrieve results from the task graph, which are keyed by experiment name.
With a little bit of data wrangling, you can now visualize the results.
And there you have it! You have successfully performed a distributed operation on your single-cell data using TileDB Cloud’s serverless compute platform. This is just the tip of the iceberg in terms of what you can do with TileDB Cloud’s task graphs but should provide a good starting point for building more complex workflows.
See the Task Graphs section of Academy to learn more.
Housekeeping
Don’t forget to clean up after yourself! You can delete the UDF you registered to avoid cluttering your TileDB Cloud workspace.