import os
import shutil
import numpy
import PIL
import tiledb
import torch
from tiledb.bioimg.converters.ome_tiff import OMETiffConverter
from tiledb.ml.readers.pytorch import PyTorchTileDBDataLoader
from tiledb.ml.readers.types import ArrayParams
root_dir = os.path.expanduser("~/tiledb-bioimg-chunked-ingestion")
if os.path.exists(root_dir):
shutil.rmtree(root_dir)
os.makedirs(root_dir)Analyze Biomedical Images with PyTorch
In this tutorial, you will delve into the practical application of deep learning techniques, specifically edge detection, to analyze biomedical images. Using the versatile PyTorch framework, you will construct and train a model capable of identifying crucial edges within these images.
To get the most out of this tutorial, some familiarity with machine learning concepts is recommended. If you’d like a refresher or deeper dive into how TileDB supports ML workflows, check out the Machine Learning section of Academy. It covers topics like storing, querying, and managing ML models efficiently, and how to integrate with other TileDB functionalities.
This tutorial requires you to run it locally and needs TileDB-Py version 0.33.5 or later and TileDB-ML version 0.9.7 or later.
Start by importing the modules you’ll need in this tutorial and creating a directory to hold the data.
Next, retrieve the image you’ll use for edge detection.
import requests
url = "https://github.com/libvips/libvips/raw/refs/heads/master/test/test-suite/images/CMU-1-Small-Region.svs"
response = requests.get(url, stream=True)
data_home = os.path.join(root_dir, "data.svs")
data_dest = os.path.join(root_dir, "data.tdb")
with open(data_home, "wb") as out_file:
shutil.copyfileobj(response.raw, out_file)Ingest the image into TileDB.
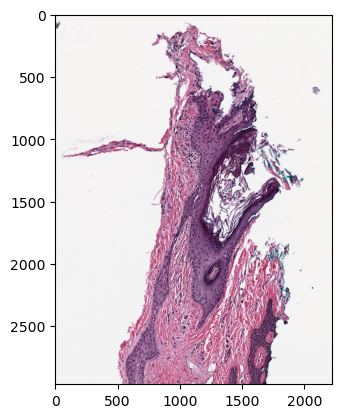
Now inspect the image to ensure it was ingested correctly.
After that, configure PyTorch and define some variables you’ll use for the ML model.
arguments_strModel = "bsds500" # only 'bsds500' for now
torch.set_grad_enabled(
False
) # make sure to not compute gradients for computational performance
torch.backends.cudnn.enabled = (
False # make sure to use cudnn for computational performance
)
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")You can design your own edge detection model, but for this example, you will use the following open source PyTorch edge detection model.
class Network(torch.nn.Module):
def __str__(self):
return super().__str__()[0:1020]
def __init__(self):
super().__init__()
self.netVggOne = torch.nn.Sequential(
torch.nn.Conv2d(
in_channels=3, out_channels=64, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
torch.nn.Conv2d(
in_channels=64, out_channels=64, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
)
self.netVggTwo = torch.nn.Sequential(
torch.nn.MaxPool2d(kernel_size=2, stride=2),
torch.nn.Conv2d(
in_channels=64, out_channels=128, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
torch.nn.Conv2d(
in_channels=128, out_channels=128, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
)
self.netVggThr = torch.nn.Sequential(
torch.nn.MaxPool2d(kernel_size=2, stride=2),
torch.nn.Conv2d(
in_channels=128, out_channels=256, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
torch.nn.Conv2d(
in_channels=256, out_channels=256, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
torch.nn.Conv2d(
in_channels=256, out_channels=256, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
)
self.netVggFou = torch.nn.Sequential(
torch.nn.MaxPool2d(kernel_size=2, stride=2),
torch.nn.Conv2d(
in_channels=256, out_channels=512, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
torch.nn.Conv2d(
in_channels=512, out_channels=512, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
torch.nn.Conv2d(
in_channels=512, out_channels=512, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
)
self.netVggFiv = torch.nn.Sequential(
torch.nn.MaxPool2d(kernel_size=2, stride=2),
torch.nn.Conv2d(
in_channels=512, out_channels=512, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
torch.nn.Conv2d(
in_channels=512, out_channels=512, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
torch.nn.Conv2d(
in_channels=512, out_channels=512, kernel_size=3, stride=1, padding=1
),
torch.nn.ReLU(inplace=False),
)
self.netScoreOne = torch.nn.Conv2d(
in_channels=64, out_channels=1, kernel_size=1, stride=1, padding=0
)
self.netScoreTwo = torch.nn.Conv2d(
in_channels=128, out_channels=1, kernel_size=1, stride=1, padding=0
)
self.netScoreThr = torch.nn.Conv2d(
in_channels=256, out_channels=1, kernel_size=1, stride=1, padding=0
)
self.netScoreFou = torch.nn.Conv2d(
in_channels=512, out_channels=1, kernel_size=1, stride=1, padding=0
)
self.netScoreFiv = torch.nn.Conv2d(
in_channels=512, out_channels=1, kernel_size=1, stride=1, padding=0
)
self.netCombine = torch.nn.Sequential(
torch.nn.Conv2d(
in_channels=5, out_channels=1, kernel_size=1, stride=1, padding=0
),
torch.nn.Sigmoid(),
)
self.load_state_dict(
{
strKey.replace("module", "net"): tenWeight
for strKey, tenWeight in torch.hub.load_state_dict_from_url(
url="http://content.sniklaus.com/github/pytorch-hed/network-"
+ arguments_strModel
+ ".pytorch",
file_name="hed-" + arguments_strModel,
).items()
}
)
# end
def forward(self, tenInput):
tenInput = tenInput * 255.0
tenInput = tenInput - torch.tensor(
data=[104.00698793, 116.66876762, 122.67891434],
dtype=tenInput.dtype,
device=tenInput.device,
).view(1, 3, 1, 1)
tenVggOne = self.netVggOne(tenInput)
tenVggTwo = self.netVggTwo(tenVggOne)
tenVggThr = self.netVggThr(tenVggTwo)
tenVggFou = self.netVggFou(tenVggThr)
tenVggFiv = self.netVggFiv(tenVggFou)
tenScoreOne = self.netScoreOne(tenVggOne)
tenScoreTwo = self.netScoreTwo(tenVggTwo)
tenScoreThr = self.netScoreThr(tenVggThr)
tenScoreFou = self.netScoreFou(tenVggFou)
tenScoreFiv = self.netScoreFiv(tenVggFiv)
tenScoreOne = torch.nn.functional.interpolate(
input=tenScoreOne,
size=(tenInput.shape[2], tenInput.shape[3]),
mode="bilinear",
align_corners=False,
)
tenScoreTwo = torch.nn.functional.interpolate(
input=tenScoreTwo,
size=(tenInput.shape[2], tenInput.shape[3]),
mode="bilinear",
align_corners=False,
)
tenScoreThr = torch.nn.functional.interpolate(
input=tenScoreThr,
size=(tenInput.shape[2], tenInput.shape[3]),
mode="bilinear",
align_corners=False,
)
tenScoreFou = torch.nn.functional.interpolate(
input=tenScoreFou,
size=(tenInput.shape[2], tenInput.shape[3]),
mode="bilinear",
align_corners=False,
)
tenScoreFiv = torch.nn.functional.interpolate(
input=tenScoreFiv,
size=(tenInput.shape[2], tenInput.shape[3]),
mode="bilinear",
align_corners=False,
)
return self.netCombine(
torch.cat(
[tenScoreOne, tenScoreTwo, tenScoreThr, tenScoreFou, tenScoreFiv], 1
)
)You can store your ML models alongside your biomedical images in TileDB. Check out the Machine Learning tutorials to get started.
Using TileDB’s dataloaders, you can fetch your images into the framework’s compatible input format.
batches = []
with tiledb.open(img_grp[0].uri) as x:
train_loader = PyTorchTileDBDataLoader(ArrayParams(x), batch_size=3)
for loaded_img in train_loader:
batches.append(loaded_img)
input_img = torch.cat(batches)--------------------------------------------------------------------------- AttributeError Traceback (most recent call last) Cell In[24], line 6 4 with tiledb.open(img_grp[0].uri) as x: 5 train_loader = PyTorchTileDBDataLoader(ArrayParams(x), batch_size=3) ----> 6 for loaded_img in train_loader: 7 batches.append(loaded_img) 8 input_img = torch.cat(batches) File /opt/conda/lib/python3.9/site-packages/torch/utils/data/dataloader.py:441, in DataLoader.__iter__(self) 439 return self._iterator 440 else: --> 441 return self._get_iterator() File /opt/conda/lib/python3.9/site-packages/torch/utils/data/dataloader.py:385, in DataLoader._get_iterator(self) 383 def _get_iterator(self) -> '_BaseDataLoaderIter': 384 if self.num_workers == 0: --> 385 return _SingleProcessDataLoaderIter(self) 386 else: 387 self.check_worker_number_rationality() File /opt/conda/lib/python3.9/site-packages/torch/utils/data/dataloader.py:672, in _SingleProcessDataLoaderIter.__init__(self, loader) 668 if isinstance(self._dataset, (IterDataPipe, MapDataPipe)): 669 # For BC, use default SHARDING_PRIORITIES 670 torch.utils.data.graph_settings.apply_sharding(self._dataset, self._world_size, self._rank) --> 672 self._dataset_fetcher = _DatasetKind.create_fetcher( 673 self._dataset_kind, self._dataset, self._auto_collation, self._collate_fn, self._drop_last) File /opt/conda/lib/python3.9/site-packages/torch/utils/data/dataloader.py:79, in _DatasetKind.create_fetcher(kind, dataset, auto_collation, collate_fn, drop_last) 77 return _utils.fetch._MapDatasetFetcher(dataset, auto_collation, collate_fn, drop_last) 78 else: ---> 79 return _utils.fetch._IterableDatasetFetcher(dataset, auto_collation, collate_fn, drop_last) File /opt/conda/lib/python3.9/site-packages/torch/utils/data/_utils/fetch.py:21, in _IterableDatasetFetcher.__init__(self, dataset, auto_collation, collate_fn, drop_last) 19 def __init__(self, dataset, auto_collation, collate_fn, drop_last): 20 super().__init__(dataset, auto_collation, collate_fn, drop_last) ---> 21 self.dataset_iter = iter(dataset) 22 self.ended = False File /opt/conda/lib/python3.9/site-packages/torch/utils/data/datapipes/_hook_iterator.py:230, in hook_iterator.<locals>.wrap_iter(*args, **kwargs) 228 @functools.wraps(func) 229 def wrap_iter(*args, **kwargs): --> 230 iter_ret = func(*args, **kwargs) 231 datapipe = args[0] 232 datapipe._snapshot_state = _SnapshotState.Iterating File /opt/conda/lib/python3.9/site-packages/torch/utils/data/datapipes/datapipe.py:364, in _IterDataPipeSerializationWrapper.__iter__(self) 363 def __iter__(self) -> "_IterDataPipeSerializationWrapper": --> 364 self._datapipe_iter = iter(self._datapipe) 365 return self File /opt/conda/lib/python3.9/site-packages/torch/utils/data/datapipes/_hook_iterator.py:230, in hook_iterator.<locals>.wrap_iter(*args, **kwargs) 228 @functools.wraps(func) 229 def wrap_iter(*args, **kwargs): --> 230 iter_ret = func(*args, **kwargs) 231 datapipe = args[0] 232 datapipe._snapshot_state = _SnapshotState.Iterating File /opt/conda/lib/python3.9/site-packages/tiledb/ml/readers/pytorch.py:104, in DeferredIterableIterDataPipe.__iter__(self) 103 def __iter__(self) -> Iterator[Any]: --> 104 return self._callable() File /opt/conda/lib/python3.9/site-packages/tiledb/ml/readers/pytorch.py:146, in _unbatch_tensors(schema, key_range) 136 def _unbatch_tensors( 137 schema: TensorSchema[TensorLike], key_range: InclusiveRange[Any, int] 138 ) -> Iterator[TensorLikeOrTuple]: 139 """ 140 Generate batches of `TensorLike`s for the given schema and key range and then unbatch 141 them into single "rows". 142 If `schema.num_fields == 1`, each "row" is a single `TensorLike` 143 If `schema.num_fields > 1`, each "row" is a sequence of `TensorLike`s 144 """ 145 batches = schema.iter_tensors( --> 146 key_range.partition_by_weight(schema.max_partition_weight) 147 ) 148 if schema.num_fields > 1: 149 # convert batches of columns to batches of rows 150 batches = (zip(*batch) for batch in batches) File /opt/conda/lib/python3.9/site-packages/tiledb/ml/readers/_tensor_schema/dense.py:69, in DenseTensorSchema.max_partition_weight(self) 67 @property 68 def max_partition_weight(self) -> int: ---> 69 memory_budget = int(self._array._ctx_().config()["sm.mem.total_budget"]) 71 # The memory budget should be large enough to read the cells of the largest field 72 bytes_per_cell = max(dtype.itemsize for dtype in self.field_dtypes) AttributeError: 'DenseArray' object has no attribute '_ctx_'
The estimation function will forward the input data into the model.
def estimate(network, input_img):
netNetwork = network.to(device).eval()
input_img = input_img.to(torch.float) * (1.0 / 255.0)
# The following values should change following the
# schema of the image if 3D CYX then accesses indices accordingly
intWidth = input_img.shape[2]
intHeight = input_img.shape[1]
return netNetwork(input_img.view(1, 3, intHeight, intWidth))[0, :, :, :]
netNetwork = Network()
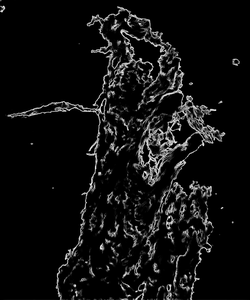
edge_detection = estimate(netNetwork, input_img)After the model finishes running on the data, you can render the output and visualize the results of the edge detection model:
Clean up in the end by removing the data directory.